Key Concepts
A DNA–protein complex in the nucleus of eukaryotic cells. Chromosomes are linear (usually) or circular structures containing deoxyribonucleic acid (DNA) complexed with histone and nonhistone proteins, a centromere, and a telomere at each end, if linear. These threadlike, tightly packed structures are seen in animals, plants, and other eukaryotes during mitotic and meiotic cell divisions (Fig. 1). The single DNA molecule in each chromosome carries a unique complement of linearly arranged genes. Chromosomes were so named because, after staining, nineteenth-century light microscopists saw chromosomes as colored bodies in cells. The combination of DNA and proteins in chromosomes is called chromatin. Collectively, the DNA from all the chromosomes in a nucleus is the hereditary blueprint for the species. Eukaryotes (plants, animals, fungi, and protists) usually have one nucleus in each cell. A nucleus is a double membrane-bound compartment in which chromosomes are located in interphase between cell divisions. In contrast, prokaryotes, such as bacteria and their allies [including mitochondria and chloroplasts (cell plastids)], do not have true chromosomes or nuclei because prokaryotes do not confine their small, single circle of DNA in a membrane-bound compartment. See also: Cell division; Deoxyribonucleic acid (DNA); Eukaryota; Gene; Histone; Meiosis; Mitosis; Protein
Interphase chromosomes
Chromosomes are usually described when they are easily observed in a condensed state during mitosis (M phase of the cell cycle). However, chromosomes are not visible during interphase, which comprises the largest part of the cell cycle. Interphase consists of G1 (the first gap in interphase without DNA synthesis from the end of telophase to the beginning of DNA synthesis), followed by S (the synthesis phase, during which the DNA is replicated, so each chromosome has two identical chromatids), and finally G2 (the second gap phase, beginning after S and lasting until M phase begins). Nuclei in cells that stop dividing usually arrest in G0, which is a G1 interphase-like state that may persist indefinitely.
During interphase, chromosomes are decondensed to form an apparently homogeneous mass of chromatin that consists of long DNA double helices associated with proteins. Chromosomes are still present as individual units in interphase nuclei, but their boundaries are difficult to distinguish (something like looking at a clear bottle full of cotton balls and trying to tell where one cotton ball ends and another begins). See also: Cell cycle
Structure and composition of mitotic chromosomes
Each chromatid of a chromosome consists of one long, linear DNA double helix that is associated with proteins to form chromatin. A DNA double helix is composed of two parallel polymeric chains of nucleotides that are twisted around each other. Each strand consists of alternating phosphate and sugar molecules with bases extending from each sugar. There are four bases: adenine (A), which forms hydrogen bonds with thymine (T) in the opposite DNA strand, and guanine (G), which forms hydrogen bonds with cytosine (C) in the opposite DNA strand. As a result, the two single DNA strands of polynucleotides in a double helix are complementary. This relationship is used when DNA is replicated during interphase of the cell cycle and when ribonucleic acid (RNA) is transcribed using DNA as a template. The DNA double helix of one chromatid is usually millions of base pairs (bp) long. See also: Nucleotide; Ribonucleic acid (RNA); Transcription
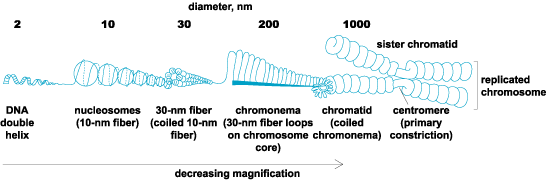
When chromosomes condense during mitosis, DNA becomes so highly folded that each double helix is approximately 10,000 times as long as the chromatid in which it is packed. (The necessity of this level of folding is apparent when one considers that diploid human cells contain about 2 m of DNA divided between 46 chromosomes in a nucleus that is about 5 μm in diameter.) In a current model for the structure of an average chromosome, the hierarchy of folding begins with formation of nucleosomes. A nucleosome consists of a core of eight basic proteins called histones (two molecules each of histones H2a, H2b, H3, and H4) and DNA. Histones are positively charged, so they are attracted to negatively charged DNA. To form a nucleosome, 146 base pairs (bp) of DNA wrap 1.65 times around the core histone particle. There are about 50 bp of coiled DNA between adjacent nucleosomes. A chain of nucleosomes linked by a continuous DNA double helix forms a fiber with a diameter of about 10 nm and a 6:1 packing ratio of DNA length to 10-nm fiber length. Next, the 10-nm fiber associates with other proteins to coil or fold into a fiber with a diameter of 30 nm and a 42:1 ratio of DNA length to 30-nm fiber length. The 30-nm fiber forms approximately 1-μm-long loops that are attached to a proteinaceous chromosome core (scaffold). The loops condense against the core under the influence of a protein complex called condensin, which is found in the core. The condensed loops and core constitute a strand called the chromonema with a diameter of about 0.2 μm. The chromonema coils, possibly due to contraction of the proteinaceous core, form a chromatid about 1 μm in diameter with a ratio of DNA length to chromatid length of approximately 10,000:1. Coiling does not occur at the primary constriction (that is, the centromere), resulting in a narrow part of the chromatid (Fig. 2). See also: Histone modifications, chromatin structure, and gene expression; Nucleosome
Without resorting to special treatments, chromosomes are visible only when they become condensed into sausage-shaped bodies during division of the nucleus (mitosis and meiosis). Although they are always microscopic, mitotic chromosomes in different species and even within a species can vary enormously in size, for example, from 1 to 40 μm in length and 1 to 2 μm in width. A chromosome can consist of either two identical sister chromatids (during G, prophase, prometaphase, and metaphase of mitosis) or one chromatid (during G1, anaphase, and telophase of mitosis). Having one or two chromatids per chromosome does not change ploidy (the number of complete chromosome sets in a nucleus). See also: Sister chromatid cohesion
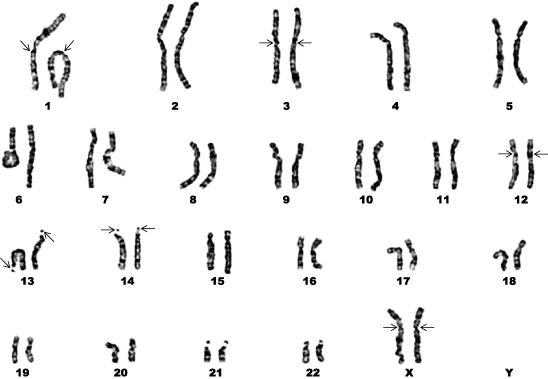
At a gross morphological level, chromosomes often can be distinguished from one another on the basis of their relative lengths and the positions of their primary constriction. The segments of chromosomes to either side of the primary constriction (centromere) are called arms. If the arms are roughly of equal length, the chromosome is metacentric. If the centromere is near one end, the chromosome is acrocentric, and there is a short (p) arm and a long (q) arm. If the centromere is truly at the end (rare), the chromosome is telocentric. Chromosomes with primary constrictions in intermediate positions are described as submetacentric and subacrocentric (Fig. 3). See also: Role of telomeres and centromeres in meiotic chromosome pairing
Kinetochores
A kinetochore is a nucleoprotein structure that forms in the centromere on each chromatid. Depending on the species, a kinetochore may appear as an amorphous sphere or a layered disk. The DNA involved in kinetochores often consists of a 150-bp tandem repeat sequence; the exact sequence varies from species to species. [In addition, there are often additional repeated sequences in the centromere and in heterochromatin (chromatin that is tightly coiled even in the nondividing nucleus and stains darkly in interphase).] Proteins found in kinetochores are more evolutionarily conserved than the DNA sequences. During mitosis, microtubules grow from the spindle poles and attach to kinetochores. Force is applied through these kinetochore microtubules to move chromosomes to the metaphase plate during prometaphase and then to move sister chromatids (now sister chromosomes) to opposite poles during anaphase.
Telomeres
The ends of chromosomes are called telomeres. Although telomeres usually have no visibly distinct structure, they are necessary for the survival of chromosomes; without telomeres, chromosomes would be nibbled away slowly from their ends during every round of DNA replication, during which a little of the DNA double helix is lost from both ends of a chromatid. The telomere is the site where an enzyme called telomerase adds expendable DNA, usually in the form of a short repeat sequence such as TTAGGG. Most differentiated animal cells have no telomerase activity, but they inherit telomeres that are thousands of base pairs long from telomerase activity during meiosis that occurred just before gametes were formed. Telomere lengths are adequate for the normal number of divisions that the cells undergo during development. This protects the body from cells that are dividing out of control because cells that divide too frequently are eventually destroyed when their telomeres are lost. One cost of this protection is aging and, as one might expect, cancer cells often have telomerase activity that permits them to divide indefinitely. See also: Aging; Cancer
Euchromatin and heterochromatin
Chromatin is divided into euchromatin and heterochromatin based on its level of condensation during interphase and G0. Euchromatin is decondensed and available for transcription. However, euchromatin still requires the proper proteins to mark genes (DNA segments) for transcription; that is, genes are not transcribed just because they are in decondensed chromatin. In contrast, heterochromatin generally remains condensed, and its DNA is not used for transcription, if for no other reason than transcription factors and RNA polymerase are sterically hindered from access to the DNA. However, even heterochromatin becomes euchromatic when its DNA is replicated during S phase. There are two types of heterochromatin: constitutive heterochromatin, which is always condensed (except during S phase), and facultative heterochromatin, which is condensed only in certain cells or in only one sex. Facultative heterochromatin seems to be a way to control gene activity.
During mitosis, euchromatin and heterochromatin are similarly condensed and stained with most dyes. However, heterochromatin in condensed chromosomes can be stained differently from euchromatin after special procedures that typically utilize Giemsa stain. One technique, called C-banding, stains constitutive heterochromatin intensely so that a simple pattern of dark bands is visible on chromosomes. C-bands often occur around centromeres, around nucleolus organizers, or at the ends of chromosomes, but they can be anywhere on a chromosome. C-banding has been used successfully on both plant and animal chromosomes. The other technique, called G-banding, stains facultative heterochromatin intensely and results in a relatively complex banding pattern. G-banding works well on mammalian and avian chromosomes and has been important for identifying individual chromosomes in these groups (Fig. 3).
Methyl groups can be added to cytosines in DNA, and this generally causes DNA to become facultatively heterochromatic and unusable for transcription. The pattern of methylation is duplicated during S phase, so methylation is inherited from one cell generation to the next. While spermatozoa and eggs carry genes for the same traits, some genes are differently methylated in spermatozoa compared with eggs. Thus, only the maternal or paternal versions of these genes are active in developing embryos. This is an epigenetic phenomenon (in which gene activity is not determined by DNA sequence, but rather through chemical modification of DNA, including methylation) called imprinting. See also: DNA methylation; Epigenome
Chromosome sets, genomes, ploidy, and homologs
Generally, eukaryotes are capable of sexual reproduction that involves fusion of two gametes. Gametes usually carry a haploid set of chromosomes, defined as one copy of each chromosome that is characteristic of the species. An unreplicated haploid set of chromosomes, in which each chromosome consists of one chromatid, contains one genome of DNA called the 1C amount (C standing for “constant” for each species). For example, the human genome (1C amount of DNA) consists of about 3.5 picograms of DNA or 3.4 billion base pairs of DNA or 1 m of DNA. Other species have different genome sizes (1C). Normally, each human gamete (spermatozoon or egg) carries a haploid chromosome set (often denoted 1n) of 23 chromosomes. After fertilization, the zygote is usually diploid (2n) with 46 chromosomes (Fig. 3 and Fig. 4). If an organism has one or more complete sets of chromosomes with no extra or missing chromosomes, it is euploid. If an organism has more than two sets, it is polyploid. If chromosomes are added or missing from a set, the organism is aneuploid. See also: Polyploidy
A special class of chromosomes called B chromosomes (or supernumerary chromosomes) occurs sporadically in many organisms. B chromosomes generally have few, if any, active genes, and they are not necessary for the life of the organism. B chromosomes may be thought of as molecular parasites, and their presence or absence does not constitute aneuploidy. Aside from B chromosomes, the numbers of chromosomes in a set vary enormously from one species to another. Generally, haploid numbers above 15 are rare in plants, and haploid numbers above 50 are rare in animals. There are two ways in which chromosome numbers can change: by breaking one chromosome into two smaller chromosomes or by fusing two chromosomes into one larger chromosome. DNA breaks are usually caused by radiation or chemicals. While breaks usually are repaired without a trace, sometimes repair leads to permanent breaks, fusion of chromosomes, trading segments between chromosomes (translocation), and inversion within a chromosome. See also: B chromosomes in plants; DNA repair
Most eukaryotic organisms are diploid with two sets of chromosomes, so each chromosome is represented twice (one derived from the egg and one from the spermatozoon). These pairs of chromosomes are called homologs or homologous chromosomes. Homologs are the same size and shape, have the same banding pattern, and carry genes for the same traits (but not necessarily the same alleles, or alternate forms, of those genes) [Fig. 3 and Fig. 4]. See also: Allele
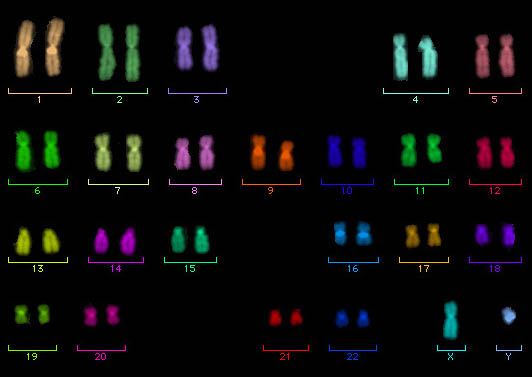
Karyotypes and idiograms
Although the terms karyotype and idiogram have sometimes been used interchangeably, it is less confusing to restrict karyotype to a word description of a set of chromosomes (for example, the haploid set of chromosomes is represented by three large metacentric, two large acrocentric, five small acrocentric, and two small metacentric chromosomes), whereas the term idiogram is used for a diagrammatic representation of a chromosome set, in which the chromosomes are arranged in a row from largest to smallest. In general, the longest chromosome is numbered 1, the next longest is numbered 2, and so on. The chromosomes can be drawn, but usually they are cut out of a photograph of a chromosome spread and pasted in position. If the cell was diploid, homologs are positioned together in pairs (Fig. 3 and Fig. 4). See also: Chromosome "fusions" in karyotype evolution
Techniques for spreading and staining chromosomes
To make chromosome preparations suitable for observation by light microscopy, cells are usually fixed (killed and preserved) in a freshly prepared solution of one part acetic acid and three parts ethanol or methanol. The chromosomes are then spread and flattened onto a glass microscope slide, either by squashing cells under a cover glass or by bursting cells that have been hypotonically swollen and permitting the chromosomes to dry on the slide. Chromosomes can then be stained for examination by light microscopy.
Many stains have been used for chromosomes. Without special treatments, dyes such as crystal violet, Giemsa stain, orcein, and carmine stain chromosomes homogeneously throughout their length. Historically, the Feulgen technique has been important because the intensity of staining is a direct indicator of the amount of DNA present. Some techniques result in the formation of “bands” on chromosomes due to differential staining based on the location of euchromatin and heterochromatin or on differences in base composition of DNA along the length of chromosomes. The latter type of banding is observed with fluorescent dyes such as Hoechst 33258, daunomycin, chromomycin A, mithramycin, DAPI, and quinacrine dihydrochloride, which either bind better or fluoresce more strongly in AT-rich or GC-rich DNA (Fig. 3).
Fluorescence in-situ hybridization (FISH)
One of the most important tools for cytogenetics is fluorescence in-situ hybridization (FISH). In this technique, a specific DNA probe is prepared by attaching a small molecule, such as biotin, to a DNA sequence of interest (for example, a gene). A solution containing the DNA probe is placed on a glass microscope slide with chromosome spreads and heated to denature both the probe and the chromosomal DNA; that is, the hydrogen bonds between AT and GC base pairs are broken so that DNA double helices separate into single, complementary DNA strands. When the slide and solution are cooled to a temperature appropriate for renaturation (reformation of double helices by hydrogen bond formation between complementary strands), the probe will renature with any complementary DNA sequence in the chromosomes. Next, an antibody specific for biotin that is itself attached to a small fluorescent dye molecule (such as fluorescein isothiocyanate, FITC) is placed on the slide. This antibody selectively binds to the biotin in the probe and then the slide is examined under a microscope that has an ultraviolet (UV) light source and the appropriate filters for the dye. The dye will fluoresce to show the location of the probe on the chromosome (or chromosomes) [Fig. 5]. FISH has proven to be a powerful technique for locating DNA sequences on chromosomes. See also: Antibody; Biotin
Other forms of FISH include genomic painting, chromosome painting, and multicolor FISH. In genomic painting, the DNA of one species is isolated and labeled with biotin. The labeled DNA is used as a probe to identify chromosomes from that species among the chromosomes from one or more different species in a hybrid. In chromosome painting, one chromosome is isolated and its DNA is amplified by the polymerase chain reaction (a technique for amplifying specific sequences of DNA that involves copying and amplifying the complementary strands of a target DNA) and labeled with biotin; this labeled DNA is then used as a probe to locate a specific chromosome in a group of chromosomes. In multicolor FISH, probes can be prepared for each chromosome in a set, and every chromosome can be identified simultaneously using different dye combinations. See also: Gene amplification; Polymerase chain reaction (PCR)
Immunofluorescence
Immunofluorescence can be used to locate specific proteins on chromosomes. The first step is the preparation of a primary antibody that will specifically bind to the protein of interest in chromosomes on a slide. Next, the primary antibody is bound by a secondary antibody that is labeled with a fluorescent dye that is visualized using fluorescence microscopy. Immunofluorescence is useful for relating proteins to the structure and function of chromosomes (Fig. 4). See also: Immunofluorescence
Function
The nucleus is a molecular library of instructions for building an organism, responding to the environment, and reproducing. Chromosomes are like molecular books, and DNA is the molecular text. In a library, most of the books and the vast majority of the text are stored compactly when not in use, but they must be available when needed. Decondensation of euchromatin in interphase makes the DNA physically available for interaction with proteins called transcription factors that recognize specific base sequences and prepare the DNA for transcription by the enzyme RNA polymerase. During transcription, complementary single-stranded RNA is made using one of the DNA strands as a template. Transcribed segments of DNA are called genes. Many RNAs are messages (messenger RNAs, or mRNAs) that are transferred to the cytoplasm, where they associate with ribosomes. The base sequence of mRNA is then used to determine the amino acid sequence of proteins in a process known as translation of the genetic code. During translation, the linear sequence of bases in mRNA is used to determine the linear sequence of amino acids in a protein. See also: Genetic code
Other RNAs are transcribed that are not messages, but instead function as structural components, enzymes, suppressors of gene activity, or signals in the pathway to forming heterochromatin. Four of these other RNAs are components of ribosomes. Most ribosomal RNA is transcribed from nucleolus organizer regions (NORs), where hundreds or thousands of copies of the ribosomal RNA gene are located. Active NORs appear as secondary constrictions on certain chromosomes (that is, chromosome constrictions in addition to centromeres). See also: Ribosomes
During mitosis, when the duplicated DNA is being divided equally between the daughter cells, the DNA is so compactly packaged in condensed chromosomes that it is temporarily unavailable for transcription, like books that are crated for shipping to another library. At the end of mitosis (telophase), chromosomes decondense and return to their interphase condition; that is, the books are uncrated, and the library is open for business.
Special chromosomes
Special chromosomes include sex chromosomes, polytene chromosomes, and the chromosomes of meiosis.
Sex chromosomes
Some animals determine whether embryos develop as males or females by the combination of sex chromosomes that they carry. Mammals and birds are prominent examples. See also: Sex chromosomes; Sex determination
In mammals, males normally have only one large X chromosome and one Y chromosome that is usually small and heterochromatic. The Y carries the SRY gene, which determines that testes will be formed rather than ovaries. Females have two X chromosomes and no Y chromosome, so they form ovaries by default (Fig. 3 and Fig. 4). However, because the number of X chromosomes differs in males and females, there is a problem with the dosage of genes on the X chromosome. Either the male with only one X chromosome has half enough genes or the female with two X chromosomes has twice too many genes. The problem is solved in mammals by converting one of the two X chromosomes in each cell of females into facultative heterochromatin in a process known as lyonization. This involves transcription of a gene on the mostly inactive X to produce RNA molecules that coat and play a role in inactivating the X chromosome. As a result, females, like males, have only one active (euchromatic) X chromosome in each cell. X inactivation occurs early in embryonic development, appears to be random with respect to which X chromosome is inactivated, and is inherited in subsequent cell divisions. Thus, the body of female mammals is a patchwork (mosaic) of tissues in which genes from one or the other X chromosome are active. During interphase, the inactive X chromosome in the nuclei of female cells stains darkly and is called a Barr body. Male cells seldom show Barr bodies. See also: Mammalia
In birds, the system is reversed: females have one Z chromosome and one small W chromosome, whereas males have two Z chromosomes. Presumably, the presence of the W chromosome somehow determines that ovaries rather than testes will form in females. Barr bodies are not visible in male birds, indicating that birds deal with dosage compensation differently than mammals. See also: Aves; Dosage compensation of the active X chromosome
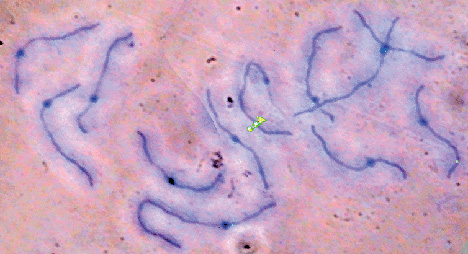
Polytene chromosomes
A few insects, plants, and protozoa have cells that undergo endoreduplication, a process involving round after round of DNA synthesis without nuclear division to produce polytene chromosomes. The best-known example of polytene chromosomes occurs in dipteran (fly) salivary glands. A polytene chromosome often consists of a thousand or more identical DNA double helices lying parallel in a bundle. Polytene chromosomes show intricate patterns of banding; in some cases, a band corresponds to a specific gene. Often euchromatin is selectively replicated, so heterochromatin has little representation. Polytene chromosomes are not comparable to mitotic chromosomes; rather, they are a special class of interphase chromosomes. Aside from protozoa that produce polytene chromosomes on the way to forming macronuclei, cells with polytene chromosomes are differentiated cells that will never divide again (Fig. 6).
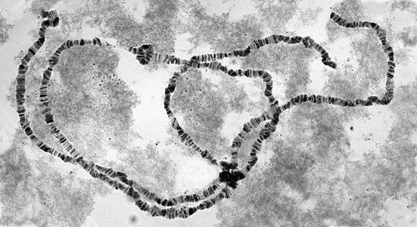
Chromosomes of meiosis
Meiosis consists of two special cell divisions referred to as meiosis I (MI) and meiosis II (MII). MI is unique among nuclear divisions because crossing over occurs between homologous chromosomes and the chromosome number is reduced from diploid to haploid. For this, elongate homologous chromosomes synapse (come together side-by-side) in a gene-for-gene alignment during prophase I. Synapsed homologs are held together by a proteinaceous structure called the synaptonemal complex (SC) [Fig. 7]. The synaptonemal complex looks like a railroad track, with the rails represented by two lateral elements. One lateral element is associated with the two sister chromatids of one chromosome, and the other lateral element is associated with the two sister chromatids of the homologous chromosome. Chromatids in homologs are called nonsister chromatids. Thin transverse filaments span the central region between the lateral elements. It is at this time that new gene combinations are created by crossing over. Pairs of homologous chromosomes held together by synaptonemal complexes or chiasmata are called bivalents. See also: Crossing-over (genetics)
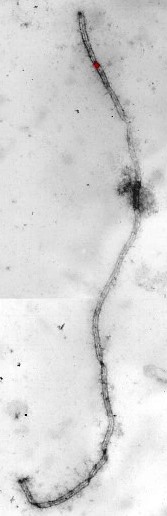
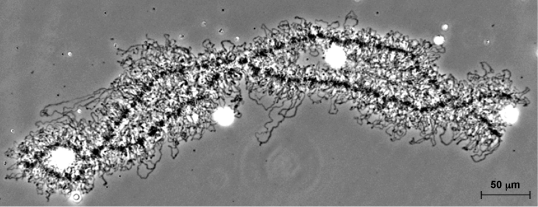
Crossing over involves breakage and repair of DNA in such a way that nonsister chromatids exchange equivalent segments. These events probably occur in recombination nodules, which are proteinaceous ellipsoids about 100 nm in diameter that reside between the lateral elements of the synaptonemal complex (Fig. 7). The pattern of crossing over (recombination) can be mapped on synaptonemal complexes by examining synaptonemal complexes and recombination nodules using electron microscopy or by locating recombination nodules using immunofluorescent antibodies targeted to a protein in recombination nodules called MLH1 (Fig. 8). Recombination nodule maps show the pattern and frequency of crossing over on chromosomes and thereby link genetic linkage maps to chromosome structure. See also: Linkage (genetics); Recombination (genetics)
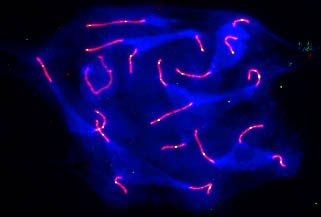
Primary oocytes (immature eggs) of most animals enter a synthetic period right after crossing over is completed and the synaptonemal complex breaks down. During this time, many components that will be needed by the developing embryo are synthesized and stored in the oocyte. Apparently to enhance transcription at this time, the pairs of homologous chromosomes (held together at sites of crossing over called chiasmata) become extremely long to permit rapid RNA synthesis on loops of chromatin extending from the chromosome axes. These bivalents (pairs of homologous chromosomes) are called lampbrush chromosomes because nineteenth-century cytologists thought that they looked like the brushes used to clean glass chimneys of oil lamps (Fig. 9).