Key Concepts
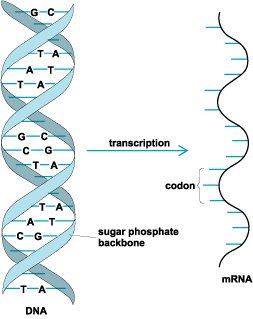
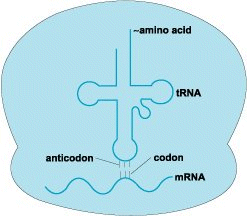
The rules by which the base sequences of DNA are translated into the amino acid sequences of proteins. The genetic code is essential for translating the information encoded within genetic material into proteins. Each sequence of deoxyribonucleic acid (DNA) that codes for a protein is transcribed or copied (Fig. 1) into messenger ribonucleic acid (mRNA). Following the rules of the genetic code, discrete elements in the mRNA, known as codons, specify each of the 20 different amino acids that are the constituents of proteins. In a process called translation, the cell decodes the message in mRNA. During translation (Fig. 2), another class of RNAs, called transfer RNAs (tRNAs), are coupled to amino acids, bind to the mRNA, and in a step-by-step fashion provide the amino acids that are linked together in the order indicated by the mRNA sequence. The specific attachment of each amino acid to the appropriate tRNA and the precise pairing of tRNAs via their anticodons to the correct codons in the mRNA form the basis of the genetic code. See also: Amino acid; Deoxyribonucleic acid (DNA); Gene; Genetics; Nucleic acid; Protein; Ribonucleic acid (RNA)
Universal genetic code
The genetic information in DNA is found in the sequence or order of four bases that are linked together to form each strand of the two-stranded DNA molecule. The bases of DNA are adenine, guanine, thymine, and cytosine, which are abbreviated A, G, T, and C, respectively. Chemically, A and G are purines, and C and T are pyrimidines. The two strands of DNA are wound about each other in a double helix that looks like a twisted ladder (Fig. 1). The two posts of the ladder consist of sugar (deoxyribose) and phosphate chains that link the bases together in a directional manner. In DNA, the two strands are said to be antiparallel because they run in opposite directions. Each rung of the ladder is formed by two bases (one from each strand) that pair with each other by means of hydrogen bonds. For a good fit, a pyrimidine must pair with a purine; in DNA, A bonds with T, and G bonds with C. See also: Hydrogen bond; Purine; Pyrimidine
Ribonucleic acids, such as mRNA or tRNA, also comprise four bases, except that, in RNA, the pyrimidine uracil (U) replaces thymine. During transcription, a single-stranded mRNA copy of one strand of the DNA is made. That mRNA copy of the DNA is bound to a large particle known as the ribosome and decoded according to the rules of the genetic code (Fig. 1 and Fig. 2). See also: Ribosomes
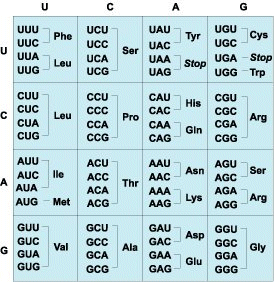
If two bases at a time are grouped together, then only 4 × 4 or 16 different combinations are possible, which is a number that is insufficient to code for all 20 amino acids that are found in proteins. However, if the four bases are grouped together in threes, then there are 4 × 4 × 4 or 64 different combinations. Read sequentially without overlapping, those groups of three bases constitute a codon, that is, the unit that codes for a single amino acid.
The 64 codons can be divided into 16 families of four (Fig. 3), in which each codon begins with the same two bases. With the number of codons exceeding the number of amino acids, several codons can code for the same amino acid. Thus, the code is degenerate. In eight instances, all four codons in a family specify the same amino acid. In the remaining families, the two codons that end with the pyrimidines U and C often specify one amino acid, whereas the two codons that end with the purines A and G specify another. Furthermore, three of the codons—UAA, UAG, and UGA—do not code for any amino acid, but instead signal the end of the protein chain.
Decoding mRNAs with tRNAs
The nucleic acid code of an mRNA is converted into an amino acid sequence with the aid of tRNAs. These RNAs are relatively small nucleic acids that vary from 75 to 93 bases in length. Some of the bases in a tRNA pair with each other, so a tRNA is folded into a pattern that can be represented in two dimensions as a cloverleaf (Fig. 2). This pattern is further folded in three dimensions to form an L-shaped molecule (Fig. 4), to which an amino acid can then be attached. At the other end, three bases in a single-stranded loop are free to pair with a codon in the mRNA. These three bases of a tRNA constitute the anticodon.
Each amino acid has one or more tRNAs. Moreover, because of the degeneracy of the code, many of the tRNAs for a specific amino acid have different anticodon sequences. However, the tRNAs for one amino acid are capable of pairing their anticodons only with the codon or codons in the mRNA that specify that amino acid. The tRNAs act as interpreters of the code, providing the correct amino acid in response to each codon by virtue of precise codon–anticodon pairing. The tRNAs pair with the codons and sequentially insert their amino acids in the exact order specified by the sequence of codons in the mRNA. This process takes place in the ribosome.
Codon–anticodon interaction
The anticodon of the tRNA pairs with the codon in the mRNA in an antiparallel fashion. Therefore, the first base of the codon pairs with the third base of the anticodon, the second base of the codon pairs with the second base of the anticodon, and the third base of the codon pairs with the first base of the anticodon. For the first two bases of the codon, pairing follows the usual rule: A pairs with U, and G pairs with C. However, at the third base of the codon, pairing occurs according to a set of rules known as the wobble hypothesis (see table).
| First base of anticodon | Third base of codon |
|---|---|
| C | G |
| U | A or G |
| G | C or U |
| I | U, C, or A |
The purine base inosine (I) is often found instead of A in the first position of the anticodon. At that location, inosine can pair with any of three bases (U, C, or A) in the third position of the codon, thus allowing a single tRNA to read three out of the four codons for a single amino acid. However, inosine is not found in tRNAs in which it would allow pairing with a codon that specifies another amino acid. For example, the histidine tRNA does not contain inosine in the first position of the anticodon because that would allow pairing with the CAA codon, which codes for glutamine.
Variations
The rules of the genetic code are virtually the same for all organisms, but there are some interesting exceptions. In the microorganism Mycoplasma capricolum, UGA is not a stop codon; instead, it codes for tryptophan. This alteration in the code is also found in the mitochondria of some organisms. In contrast, UGA is the sole termination codon in some ciliated protozoa, where UAA and UAG are translated as glutamine. Other specific changes in the meaning of codons that normally encode isoleucine, leucine, and arginine have also been observed in the mitochondria of some organisms. In addition, a modified system for reading codons that requires fewer tRNAs is found in mitochondria. See also: Mitochondria; Mycoplasmas
Aminoacyl-tRNA synthetases
The specific attachment of amino acids to tRNAs is catalyzed by a class of enzymes known as aminoacyl-tRNA synthetases. Each amino acid has a different aminoacyl-tRNA synthetase, which catalyzes a reaction known as aminoacylation. Each enzyme attaches its particular amino acid to all of the tRNAs that have anticodons corresponding to that amino acid. Each aminoacyl-tRNA synthetase must distinguish between the various tRNAs so that only the proper ones are aminoacylated. Thus, the accurate recognition of tRNAs by these enzymes establishes the rules of the genetic code. See also: Enzyme
Because synthetases are responsible for the genetic code and are present in all living organisms, it is likely that they are among the oldest enzymes on Earth. The active-site region of a synthetase is thought to be the primordial, or earliest, form of the synthetase. In addition to the active-site-containing region that is conserved among all members of the same class, each synthetase has a region that is referred to as a nonconserved domain. This domain is more or less unique to the synthetase, even within a particular class.
Second genetic code
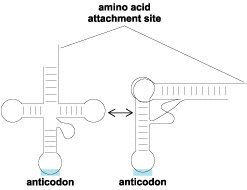
In the case of the tRNA specific for alanine with an anticodon GGC, a single G–U base pair, at a specific position far removed from the anticodon, is needed for aminoacylation with alanine. This base pair is located in a portion of the tRNA molecule relatively close to the site of amino acid attachment. The part of the tRNA structure that is near the amino acid attachment site is known as the acceptor stem, and it consists of a short helix joined to a single-stranded piece that terminates at the site of amino acid attachment. A truncated version of alanine tRNA that contains the critical base pair together with the adjacent structure near the amino acid attachment site can be synthesized as a minihelix. This tRNA fragment is recognized by alanine tRNA synthetase.
Even smaller RNA fragments that recapitulate the acceptor ends of several tRNAs are aminoacylated with the correct amino acid. Because aminoacylation is specific and depends on the order of bases in the short fragments, the relationship between the structure-sequence of RNA fragments and each amino acid can be viewed as an operational RNA code, often referred to as the second genetic code.
Several synthetases recognize the anticodon as one of the elements needed for identification of the tRNA; however, even in these cases, nucleotides in the acceptor stem are still important for precise and efficient recognition of the whole tRNA and small RNA fragments. The class-defining and active-site-containing domain of aminoacyl-tRNA synthetases is largely responsible for interactions with the acceptor end of a tRNA and, therefore, for interpretation of the second genetic code. After establishment of the second genetic code in a primitive world, RNA fragments with signals for attachment of specific amino acids may have become incorporated into RNA structures that eventually became transfer RNAs. That step, in turn, gave birth to the modern genetic code. In addition, the synthetases developed editing functions that are tRNA-dependent. The development of these editing functions was one of the events that linked the evolution of tRNAs with that of the synthetases. These editing functions enable the synthetases to achieve high accuracy of aminoacylation. During evolution, the earliest tRNA synthetases apparently acquired additional, unique domains that allowed for editing. See also: Nucleotide





