Key Concepts
Any of a group of viruses that cause upper and lower respiratory tract diseases, gastroenteritis, hepatitis, and central nervous system infections in birds and mammals, including humans. Coronaviruses (Fig. 1) comprise a major group of common animal viruses that typically infect mammals and birds. Most notably, since the start of the twenty-first century, coronaviruses have emerged as threatening zoonotic pathogens (originating in wild animals) that have caused three large-scale pandemics: severe acute respiratory syndrome (SARS), Middle East respiratory syndrome (MERS), and coronavirus disease 2019 (COVID-19). The COVID-19 pandemic is a once-in-a-century event similar in scope to the influenza pandemic of 1918–1919, and these two pandemics have caused high morbidity, mortality, and social and economic disruption on a global scale. Mild, less-severe coronavirus diseases of the upper respiratory tract include certain cases of the common cold. More-severe coronavirus infections can lead to lower respiratory illness (pneumonia), gastroenteritis, peritonitis, reproductive diseases, nephritis (kidney disease), hepatitis, and central nervous system infections. See also: Animal virus; Detection of respiratory viruses; Epidemic; Epidemiology; Infectious disease; Middle East respiratory syndrome (MERS); Novel coronavirus is declared a global pandemic; Pathogen; Respiratory system disorders; Severe acute respiratory syndrome (SARS); Virus; Zoonoses
Background
Observations of coronavirus infections were first reported in the United States in 1931. Infected chickens on a North Dakota farm were listless and gasping for air, resulting in high mortality. Infected embryos were born dwarfed, and their joints were fused. It was determined that the "gasping disease"—infectious bronchitis—was caused by a filterable agent, that is, infectious bronchitis virus (IBV). During the 1960s, the first human coronaviruses were isolated from the nasal washings of a child who had the typical signs and symptoms of a common cold. See also: Common cold
Prior to the SARS pandemic in 2003, human coronavirus 229E (HCoV 229E) and HCoV OC43 were the only known coronavirus strains circulating in the human population, causing 15–29% of all common colds. Research indicates that more than 30% of children test positive for antibodies (evidence of infection) with either coronavirus strain within the first 12 months of life. See also: Antibody
Coronavirus structure
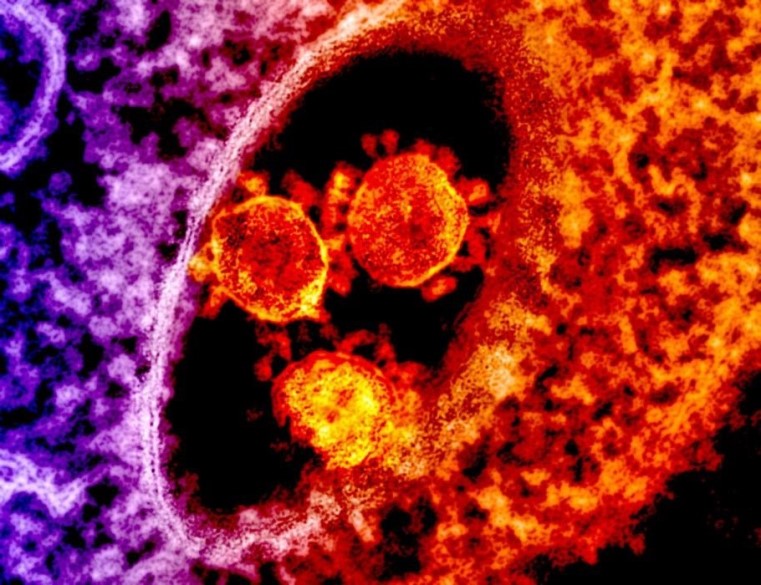
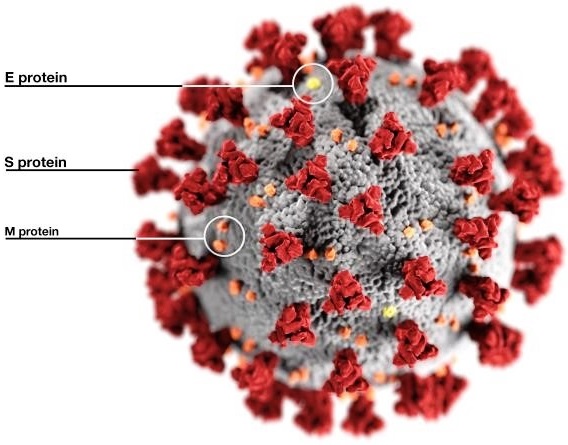
The name coronavirus is derived from the Latin word corona because its spike (S) proteins (Fig. 2), which protrude outside of the virion (the complete, mature virus particle), resemble a royal crown or the Sun’s corona when viewed using an electron microscope (Fig. 1). Coronaviruses are enveloped and contain a single-stranded ribonucleic acid (RNA) genome. The S protein is responsible for the attachment of the virus to a receptor present on the surface of a host cell, and the protein mediates the fusion of the coronavirus envelope with cellular membranes, allowing entry into the host cell. In addition, the S protein is the main target for antibodies produced by the body as an immune defense against the coronavirus. The envelope is associated with two additional transmembrane proteins: a small envelope (E) protein and a membrane (M) protein (Fig. 2). Some coronaviruses contain an additional envelope protein, which is a hemagglutinin-esterase (HE) spike protein. See also: Cell membrane; Genomics; Immunology; Ribonucleic acid (RNA)
Coronavirus genomes range in length from 26 to 32 kilobases. These genomes are the longest in size of all viruses containing RNA genomes. Similar to most eukaryotic messenger RNAs, coronavirus genomic RNA contains a 5′-end cap structure and a polyadenylated 3′-end. Table 1 and Table 2 list various characteristics of the coronaviruses that infect humans.
| HCoV 229E | HCoV OC43 | HCoV HKU1 | HCoV NL63 | |
|---|---|---|---|---|
| Year isolated | 1962 | 1965 | 2004 | 2004 |
| Classification | Alphacoronavirus | Betacoronavirus | Betacoronavirus | Alphacoronavirus |
| Receptor | Aminopeptidase N (APN) | Sialic acid | Sialic acid | Angiotensin-converting enzyme 2 (ACE-2) |
| Incubation period | 2–5 days | 2–5 days | 2–4 days | 2–4 days |
| Mode of transmission | Respiratory droplets, contaminated fomites | Respiratory droplets, contaminated fomites | Respiratory droplets, contaminated fomites | Respiratory droplets, contaminated fomites |
| Disease symptoms | Malaise, headache, nasal discharge, sneezing, sore throat, fever, and cough (common cold); pneumonia in immune-compromised patients | Malaise, headache, nasal discharge, sneezing, sore throat, fever; and cough (common cold); occasionally associated with pneumonia | Fever, runny nose, cough, and dyspnea (shortness of breath); occasionally associated with pneumonia in infants, elderly, and immune-compromised patients; gastrointestinal symptoms have been reported | Cough, croup, runny nose, rapid and shallow breathing, and hypoxia; associated with obstructive laryngitis in children |
| Epidemiology | Cases occur globally; peaks in winter | Cases occur globally; peaks in winter | Cases occur globally; peaks in winter | Cases occur globally; peaks in winter |
| Reservoir (primary host) | Bats | Rodents | Rodents | Bats |
| Intermediate host | Alpaca? | Cattle? | ? | ? |
| SARS-CoV-1 | MERS-CoV | SARS-CoV-2 | |
|---|---|---|---|
| Year isolated | 2003 | 2012 | 2019 |
| Classification | Betacoronavirus | Betacoronavirus | Betacoronavirus |
| Receptor | Angiotensin-converting enzyme 2 (ACE-2) | Dipeptidyl peptidase-4 (DPP4) [also known as CD26] | Angiotensin-converting enzyme 2 (ACE-2) |
| Alternative receptor(s)? | Dendritic cell-specific ICAM-3-grabbing non-integrin (DC-SIGN); DC-SIGN-related protein (DC-SIGNR); liver and lymph node sinusoidal endothelial cell C-type lectin (LSECtin) | ? | ? |
| Incubation period | 2–14 days (mean: 5 days) | 2–14 days (mean: 4–5 days) | 2–14 days (mean: 5–7 days); possibly up to 20 days |
| Mode of transmission | Respiratory droplets, contaminated fomites, oral-fecal? | Respiratory droplets, contaminated fomites | Respiratory droplets, airborne, contaminated fomites, efficient human-to-human community spread, oral-fecal? |
| Mortality rate | 10% | 34% | About 3.5% |
| Disease symptoms | Fever, myalgia, headache, malaise, dry cough, dyspnea (shortness of breath), respiratory distress, and diarrhea | Fever, cough, chills, sore throat, myalgia, arthralgia, dyspnea (shortness of breath), pneumonia, diarrhea, vomiting, and acute kidney failure | Fever, dry cough, dyspnea (shortness of breath), diarrhea, respiratory distress, conjunctivitis, loss of taste and smell, and acute kidney failure (elderly high risk) |
| Epidemiology | 2002–2003 in China; pandemic spread | 2012 in Middle East; 2015 in South Korea; endemic in the Middle East | 2019–2020 in China; pandemic spread |
| Reservoir (primary host) | Bats (Rhinolophus) | Bats (Taphozous, Rhinopoma, and Pipistrellus) | Bats (Rhinolophus) |
| Intermediate host | Masked civet cats, raccoon dogs, and ferret badgers (frequently sold as food sources at live markets in China) | Dromedary camels | Malayan pangolin? |
The viral genome is coated or complexed with nucleocapsid (N) proteins to form a helical nucleocapsid. In addition to the genes that code for the structural proteins that are the building blocks of the coronavirus particle (S, HE, E, M, and N proteins), the genome codes for 16 or more additional genes that function in replication, manipulating the activities of the host cell; these activities include slowing down the cell's protein synthesis machinery, allowing the adaptation to different tissues or cell types of hosts, and evading an affected body’s immune system.
Coronavirus research
Before the SARS pandemic in 2003, coronaviruses were not known to be highly pathogenic in humans. In particular, the HCoV 229E and OC43 strains were exclusively studied by researchers because the strains could be adapted to growth in laboratory tissue cultures, thereby facilitating experimental investigations.
Mouse hepatitis virus (MHV) is one of the most studied coronaviruses. MHV was discovered in 1950 when a laboratory breeding stock of white mice died from fatal hepatitis. MHV caused the hepatitis, and researchers later determined that MHV could be isolated from paralyzed white mice suffering from encephalomyelitis [the destruction of myelin (a fatty substance that forms a sheath around certain nerve fibers) as a result of inflammation in the brain and spinal cord, which can be triggered by viral infections]. Subsequently, mice were used as a model to study fundamental questions about human hepatitis and multiple sclerosis (a demyelinating disease of the nervous system). In particular, scientists were interested in whether there were genetic factors in multiple sclerosis associated with clearing viral infections and what role the immune system played in causing the pathogenesis observed in multiple sclerosis. See also: Hepatitis; Multiple sclerosis
Other pre-SARS research focused on coronavirus infections having veterinary and economic importance. Various respiratory and gastrointestinal diseases of pigs, cattle, horses, chickens, turkeys, dogs, and cats caused by coronavirus infections were studied. For example, porcine transmissible gastroenteritis resulted in mortalities approaching 100% in newborn pigs, but there also were strains of porcine coronaviruses that caused mild respiratory illness with no gastrointestinal symptoms. In addition, symptoms of winter dysentery in dairy cattle included bloody diarrhea, respiratory illness, and low milk yields during the winter months, and bovine shipping fever (pneumonia) was noted in feedlot cattle (raised for beef). Coronavirus infections were also reported in other species of animals, including ferrets, rabbits, guinea pigs, and rats.
Since the onset of the SARS pandemic in 2003, scientific investigators sought to identify the zoonotic reservoir or source of SARS-CoV-1, and its transmission to humans. For example, researchers used serological surveillance methodologies to screen for antibodies against coronaviruses present in blood samples of domestic and wild mammals and birds (including animals on farms and those sold in live markets), leading to the discovery of a significant number of novel coronaviruses. Moreover, scientists isolated dozens of coronaviruses from a wide variety of bat species (order Chiroptera) in Asia, Africa, Europe, and America. In addition, the coronavirus that causes MERS was detected in dromedary camels. See also: Chiroptera
In 2019, coronaviruses were found in the lung samples of two dead Malayan pangolins (order Pholidota) in Guangdong province in China. Pangolins are an endangered species and are the most illegally trafficked mammal in the world. In China, they are used as a food source, and their scales are used in traditional Chinese medicine. The two aforementioned pangolins had frothy liquid in their lungs and pulmonary fibrosis (a pathology also observed in COVID-19 patients). Genetic analysis determined that the coronaviruses from these pangolins were 91% and 90.5% identical to SARS-CoV-2, which was the virus responsible for the COVID-19 outbreak that began in Wuhan, China, toward the end of 2019. These results suggest that SARS-CoV-2 may have originated in pangolins, which acted as intermediate hosts of the virus. However, other possible animal sources of SARS-CoV-2 include bats as primary hosts (reservoirs) of the virus. See also: Pholidota
Coronavirus classification/taxonomy
Coronaviruses are classified in the family Coronaviridae and the subfamily Orthocoronavirinae. The subfamily is divided into four genera, based on differences in protein sequences: Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus (Fig. 3). Alphacoronaviruses and betacoronaviruses circulate in mammals, including bats, especially causing respiratory illnesses in humans and gastroenteritis in animals. These viruses also pose a heavy burden on livestock. Deltacoronaviruses mostly infect birds, whereas gammacoronaviruses infect birds and mammals. See also: Virus classification
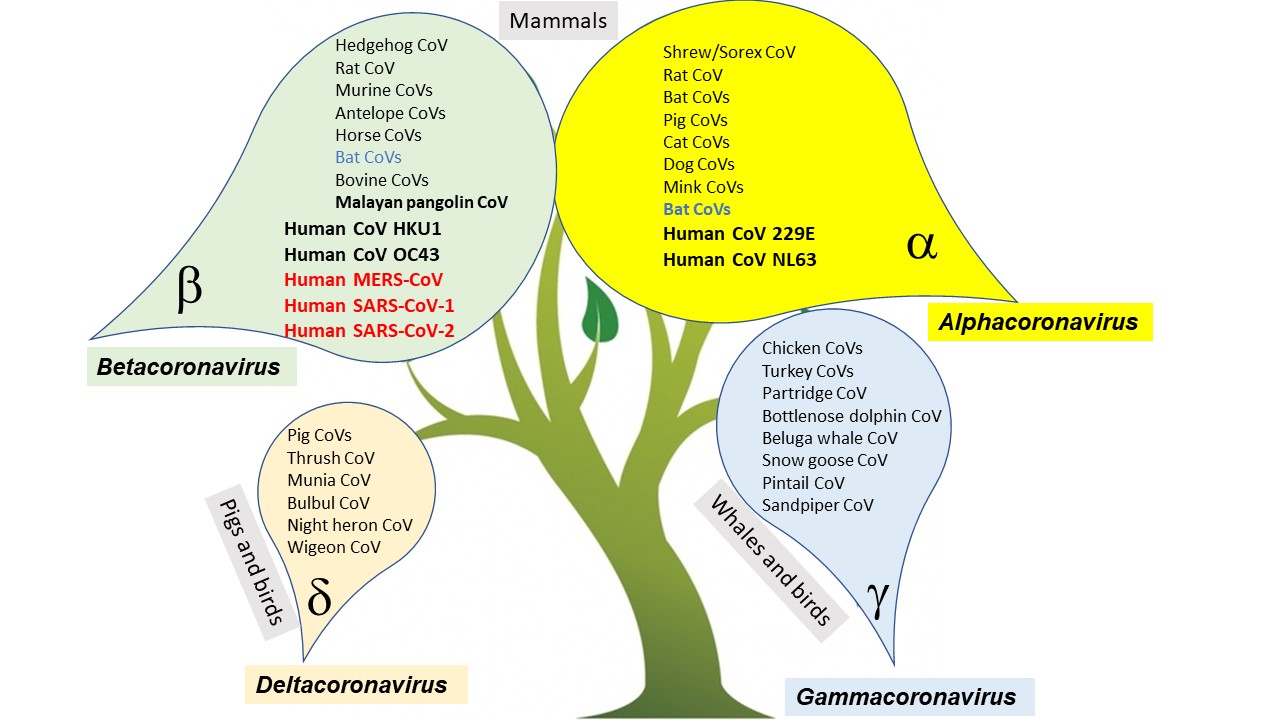
Seven coronaviruses are known to cause infections in humans. Four of these coronaviruses cause mild respiratory illness: HCoV 229E, HCoV OC43, HCoV NL63, and HCoV HKU1 (Table 1). The remaining three coronaviruses are emergent viruses that cause severe respiratory distress/pneumonia and even death: SARS-CoV-1, MERS-CoV, and SARS-CoV-2 (Table 2). All of the human coronavirus genomes have been sequenced. Based on a comprehensive comparison to the coronavirus genome sequences available in genomic databases, all human coronaviruses have animal origins.
Coronaviruses and spillover events
The term "species barrier" encompasses the concept that natural mechanisms prevent the crossing of pathogens from one species to another. The term spillover refers to the transmission of a pathogen from a host species to a recipient species (for example, from a vertebrate animal to a human). Research conducted before 2002 demonstrated that mammalian coronaviruses could cross species barriers by infecting birds. Bovine coronaviruses also could infect turkeys, but not chickens and pigs. However, the 2002 spillover of SARS-CoV-1 jumping from wild animals to humans surprised the medical community. See also: Disease ecology
The first cases of SARS in 2002 were observed in a wild-animal trader, restaurant chefs, and exotic-animal food handlers, who butchered and sold the meat at live markets in Guangdong province in China. In China, the meat of wild animals is considered more natural and nutritious compared to farmed meat. Freshly prepared meat is preferred. Live animals, typically kept in stacked open-air cages, may be bought from hundreds of small markets. The stressed animals may be infected and maintained in cramped unsanitary conditions, allowing coronaviruses (and other pathogens) to spread more easily.
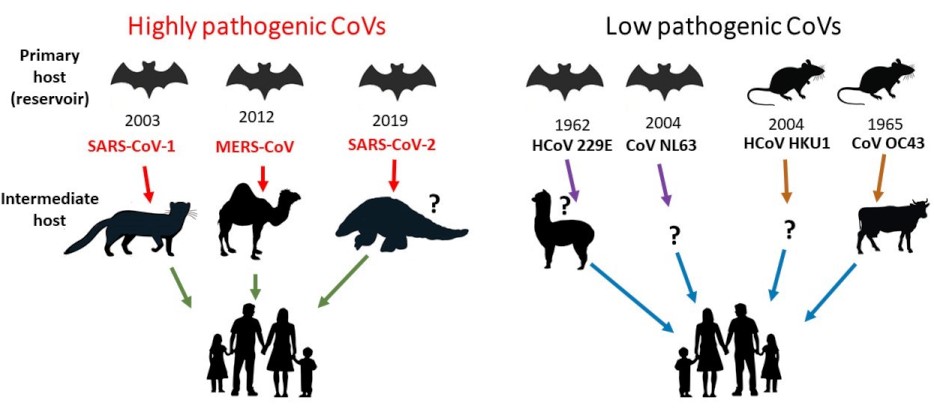
During the early SARS outbreak, scientists used molecular probes to detect certain regions of the coronavirus genome in the domestic and wild animals being sold at the live markets. Moreover, blood samples were drawn from workers with exposure to the animals, and serological surveys detected the presence of coronavirus antibodies. The animal traders and food handlers had significantly higher antibody titers against coronaviruses compared to the general population. Thus, the animals sold at the live markets were the likely source of SARS-CoV-1. The first animals found to carry SARS-CoV-1 were palm civets and raccoon dogs. After culling of the masked civets at the live markets, SARS cases ceased. Further investigations determined that Chinese horseshoe bats (Rhinolophus) were the primary host or natural animal host (reservoir) of SARS-CoV-1. The current knowledge of primary and intermediate hosts for human coronaviruses is summarized in Fig. 4.
Many wild animals have been surveyed since SARS-CoV-1 was isolated in 2003. In addition to coronaviruses, bats continue to harbor some of the most diverse and highly pathogenic viruses, such as Ebola, Nipah, Hendra, and rabies viruses. Bats are found everywhere on Earth, with the exception of Antarctica, and account for 20% of all mammals. They play an important role in agriculture by pollinating fruit trees and controlling insect populations (for example, mosquitoes). In Asia, more than 50 species of bats are hunted and eaten by low-income populations. Bats also are used in traditional medicine, and their guano (excrement) is harvested to use as fertilizer for jackfruit, eggplant, papaya, and other economically important crops.
Bats are capable of sustained flying speeds up to 160 km per hour (99 miles per hour) for hours at a time, achieving long distances that aid virus transmission. Because they possess remarkably strong antiviral immune responses and limited self-damaging inflammatory responses, bats rarely exhibit disease symptoms. Bats are being increasingly recognized as flying germ factories for zoonotic diseases that become lethal when they jump to humans. Undoubtedly, the conditions at some geographic locations will be ideal for spillover and a potential epidemic.
Coronaviruses are masters of RNA recombination
What determines whether a coronavirus will jump to humans? Interestingly, the mutation rate of SARS-CoV-1 has been estimated to be consistent with that of other viruses that have RNA genomes. The viral RNA genome has a high-fidelity rate, undergoing 0.17 mutations per genome per day or 2 mutations per human passage (few errors occur during viral replication). See also: Mutation
Coronaviruses are masters of RNA recombination, which is a genetic mechanism whereby two different viral RNA genomes hybridize and exchange regions of RNA at precise locations. Coronavirus genomes undergo RNA recombination at high frequencies during replication. In doing so, the integrity of the RNA is preserved (replacing any mistakes made during replication). In addition, coronaviruses can pick up novel genes, enabling them to adapt to a new host. RNA recombination can occur between the same and different coronavirus genomes (and between host RNA, which is less common). See also: Recombination (genetics)
The RNA region that codes for the S protein is located in the most variable region of the coronavirus genome. RNA recombination events resulting in S-protein mutations are typical examples of how viral genomic changes allow a coronavirus to adapt and attach to a host receptor, enabling the coronavirus to infect a different species. In addition, changes in other locations of the coronavirus genome can increase its adaptability to new hosts, or the genome may incorporate host/cellular RNA into its genome that codes for an accessory factor that permits its replication in a different species. Thus, it is more apparent than ever that emerging coronaviruses are a serious threat.





